Tutorial
Plotting and Clustering Megapit Soils Data
Authors: Donal O'Leary, Claire Lunch
Last Updated: Nov 23, 2020
This tutorial will show you how to download NEON Megapit soils data, plot soil profiles by texture and chemical proterties, and to cluster the different megapit profiles according to their similarity across multiple dimensions.
Objectives
After completing this activity, you will be able to:
- Download NEON megapit data using the
neonUtilitiespackage. - Join megapit data tables
- Plot profiles of megapit data by horizon
- Cluster sites into groups based on physical and chemical properties
Things You'll Need To Complete This Tutorial
To complete this tutorial you will need R (version >3.6) and, preferably, RStudio loaded on your computer.
Install R Packages
- neonUtilities: Basic functions for accessing NEON data
- dplyr: Data manipulation functions
- aqp: Algorithms for Quantitative Pedology
- cluster: Clustering utilities
- sharpshootR: Plotting tools for clustered data
- Ternary: Tools for making Ternary plots
These packages are on CRAN and can be installed by
install.packages().
Additional Resources
Load megapit data to R
Before we get the data, we need to install (if not already done) and load the R packages needed for data load and analysis.
# Install packages if you have not yet.
install.packages("neonUtilities")
install.packages("aqp")
install.packages("cluster")
install.packages("sharpshootR")
install.packages("dplyr")
install.packages("Ternary")
# Set global option to NOT convert all character variables to factors
options(stringsAsFactors=F)
# Load required packages
library(neonUtilities)
library(aqp)
library(cluster)
library(sharpshootR)
library(dplyr)
library(Ternary)
Megapit physical and chemial properties:
In this exercise, we want all available data, so we won't subset by site or date range.
MP <- loadByProduct(dpID = "DP1.00096.001", check.size = F)
# Unlist to environment - see download/explore tutorial for description
list2env(MP, .GlobalEnv)
Merge Tables
We'll join the horizon data to the physical and chemical characteristics data in order to see a depth profile of biogeochemical characteristics. The variables needed to join correctly are horizon (either name or ID) and either pitID or siteID, but we'll include several other columns that appear in both tables, to avoid creaing duplicate columns.
# duplicate the 'horizon' information into a new table
S <- mgp_perhorizon
# duplicate the biogeochemical information into a new table
B <- mgp_perbiogeosample
# Select only 'Regular' samples (not audit)
B <- B[B$biogeoSampleType=="Regular" &
!is.na(B$biogeoSampleType),]
# Join biogeochem data to horizon data
S <- left_join(S, B, by=c('horizonID', 'siteID',
'pitID','setDate',
'collectDate',
'domainID',
'horizonName'))
S <- arrange(S, siteID, horizonTopDepth)
There are two more things that we will want to do before converting this dataframe into a SoilProfileCollection object. First, we will make a new siteLabel column to use when plotting seeral pedons at once.
## combine 'domainID' and 'siteID' into a new label variable
S$siteLabel=sprintf("%s-%s", S$domainID, S$siteID)
Second, we will convert the soil physical properties (sand, silt, clay percentages) into an RGB color that we can use for plotting later. These steps are much easier to do now while the data are in a simple data.frame.
# duplicate physical property variables
S$r=S$sandTotal # Sand is Red 'r'
S$g=S$siltTotal # Silt is Green 'g'
S$b=S$clayTotal # Clay is Blue 'b'
# set 'na' values to 100 (white)
S$r[is.na(S$r)]=100
S$g[is.na(S$g)]=100
S$b[is.na(S$b)]=100
# normalize values to 1 and convert to vector of colors using 'rgb()' function
S$textureColor=rgb(red=S$r/100,
green=S$g/100,
blue=S$b/100,
alpha=1,
maxColorValue = 1)
We now have a data frame of biogeochemical data, organized
by site and horizon. We can convert this to a
SoilProfileCollection object using the aqp package.
depths(S) <- siteLabel ~ horizonTopDepth + horizonBottomDepth
We then move the site-level attributes to the @site 'slot' of S, our AQP object.
This makes our S object easy to subset to look at a particular site. (Thanks
to Dylan Beaudette of the USDA-NRCS for the helpful tips on how to best use
this package!)
site(S) <- ~ siteID + nrcsDescriptionID
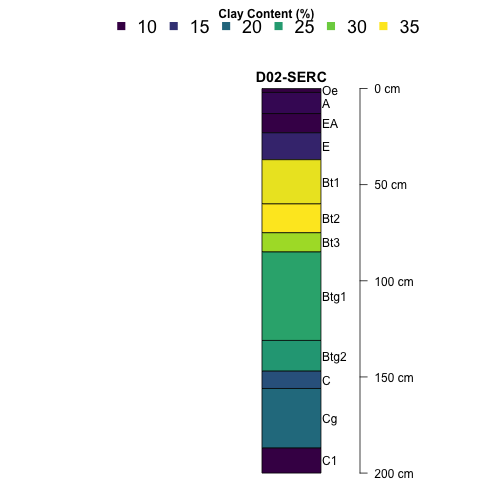
Plot Simple Soil Profiles
Using the plotting functions in the aqp package,
let's start exploring some depth profiles. We'll start with
a single site, the Smithsonian Environmental Research Center
(SERC), and plot clay content by depth.
# adjust margins
par(mar=c(1,6,3,4), mfrow=c(1,1), xpd=NA)
# Plot SERC clay profile
plotSPC(subset(S, siteID=="SERC"),
name='horizonName', label='siteLabel',
color='clayTotal', col.label='Clay Content (%)',
col.palette=viridis::viridis(10), cex.names=1,
width = .1, axis.line.offset = -6,
col.legend.cex = 1.5, n.legend=6,
x.idx.offset = 0, n=.88)
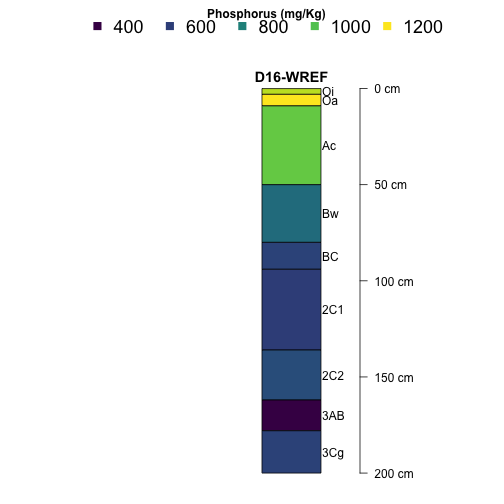
Now let's take a look at phosphorus at Wind River Experimental Forest (WREF).
# adjust margins
par(mar=c(1,6,3,4), mfrow=c(1,1), xpd=NA)
plotSPC(subset(S, siteID=="WREF"),
name='horizonName', label='siteLabel',
color='pMjelm',
col.label='Phosphorus (mg/Kg)',
col.palette=viridis::viridis(10), cex.names=1,
width = .1, axis.line.offset = -6,
col.legend.cex = 1.5, n.legend=4,
x.idx.offset = 0, n=.88)
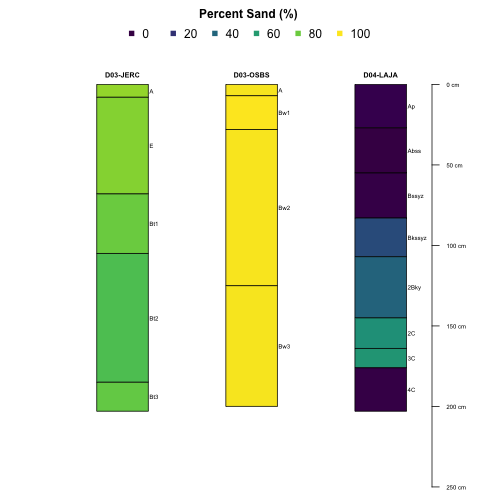
Plotting Multiple Sites
We can even pass the plotting function multiple sites in order to compare the pedons directly:
par(mar=c(0,2,3,2.5), mfrow=c(1,1), xpd=NA)
plotSPC(subset(S, siteID %in% c('JERC', 'OSBS', 'LAJA')), # pass multiple sites here
name='horizonName', label='siteLabel',
color='sandTotal',
col.label='Percent Sand (%)',
col.palette=viridis::viridis(10),
n.legend = 5)
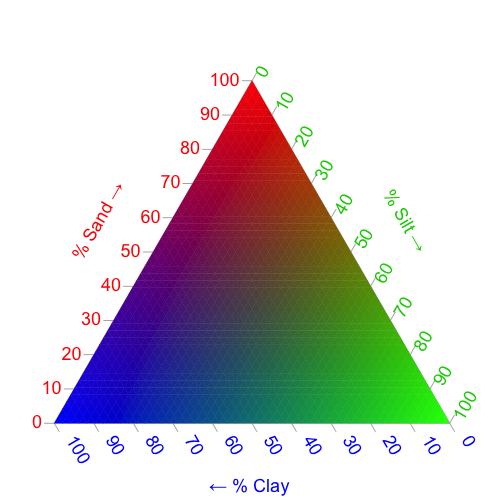
Multivariate Plotting
When analyzing soils data, we are often interested in more than one variable at a time. A classic example is texture, which in its simplest terms, is described in terms of percent Sand, Silt, and Clay. In order to plot these three variables at a time, we can describe each percentage as a color on the color wheel. Remember, in the section above, we assigned the color Red to Sand, Green to Silt, and Blue to Clay. We can plot these colors on the familiar three-axis plot that is often used to describe soil texture to serve as our color legend. To do this, we will use the Ternary package:
# Set plot margins
par(mfrow=c(1, 1), mar=rep(.3, 4))
# Make ternary plot grid
TernaryPlot(alab="% Sand \u2192", blab="% Silt \u2192", clab="\u2190 % Clay ",
lab.col=c('red', 'green3', 'blue'),
point='up', lab.cex=1.5, grid.minor.lines=1, axis.cex=1.5,
grid.lty='solid', col=rgb(0.9, 0.9, 0.9), grid.col='white',
axis.col=rgb(0.6, 0.6, 0.6), ticks.col=rgb(0.6, 0.6, 0.6),
padding=0.08)
# Define colors for the background
cols <- TernaryPointValues(rgb)
# Add colors to Ternary plot
ColourTernary(cols, spectrum = NULL, resolution=45)
Now, we can plot the pedons according to their soil texture:
par(mar=c(0,2,3,2.5), mfrow=c(1,1), xpd=NA)
plotSPC(subset(S, siteID %in% c('JERC', 'OSBS', 'LAJA')), # pass multiple sites here
name='horizonName', label='siteLabel',
color='textureColor')

Multivariate Clustering
We have 47 Megapit samples across the NEON observatory, spanning a wide range of soil types, textures, and chemical profiles. While it may be helpful from a geographic perspective to group the pedons by Site ID, it may be even more helpful to group the samples by their inherent properties. For example, grouping soils by texture, or by their organic matter content.
In order to make these groupings, we will employ a DIvisive ANAlysis (DIANA) clustering technique using the cluster package. First, let's group by soil texture:
d <- profile_compare(S, vars=c('clayTotal','sandTotal', 'siltTotal'),
k=0, max_d=100)
## Computing dissimilarity matrices from 47 profiles [1.28 Mb]
# vizualize dissimilarity matrix via divisive hierarchical clustering
d.diana <- diana(d)
# Plot the resulting dendrogram
plotProfileDendrogram(S, d.diana, scaling.factor = .6,
y.offset = 2, width=0.25, cex.names=.4,
name='horizonName', label='siteLabel',
color='textureColor')
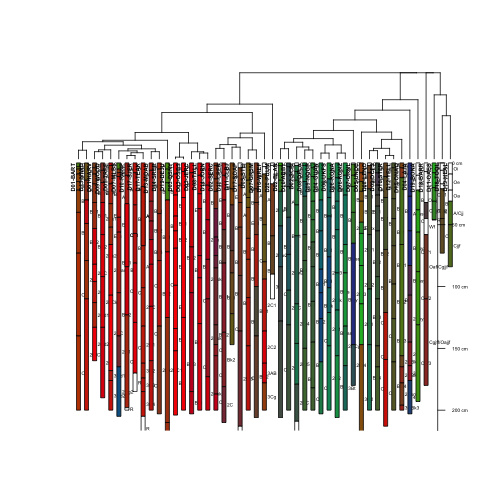
Well, it seems to have worked and plotted, but it is really hard to see all of this information in the small plotting area.
In order to visualize these data, we will need to make a plotting area large enough to contain the full plot with labels. To do so, we will open a PDF graphics device, generate the plot in that device, then close and save the device using dev.off(). It will also be a good idea to check your current working directory, and perhaps change that to where you want to save your PDFs.
# Check and set working directory as needed.
getwd()
## [1] "/Users/olearyd/Git/data"
# setwd("enter path to save PDF here")
# Open 'pdf' graphic device. Define file name and large dimensions
pdf(file="NEON_Soils_Texture_Color_Clusters.pdf", width=24, height=10)
# set plot margins and generate plot
par(mar=c(12,2,10,1), mfrow=c(1,1), xpd=NA)
plotProfileDendrogram(S, d.diana, scaling.factor = .6,
y.offset = 2, width=0.25, cex.names=.4,
name='horizonName', label='siteLabel',
color='textureColor')
# Close and save the device
dev.off()
## quartz_off_screen
## 2
This plot is not shown on this tutorial webpage, but you can view and download an example of the
Clustering by Nutrients
Rather than cluster based on physical properties, we can also cluster based on nutrient contents (nitrogen, carbon, and sulfur):
## Cluster as above, but for nutrient variables
d.nutrients <- profile_compare(S,
vars=c('nitrogenTot','carbonTot', 'sulfurTot'),
k=0, max_d=100)
## Computing dissimilarity matrices from 47 profiles [1.27 Mb]
# vizualize dissimilarity matrix via divisive hierarchical clustering
d.diana.nutrients <- diana(d.nutrients)
Let's make another PDF for our plot. However, this time is itsn't as straightforward to plot the three nutrients of interest as 'rgb' colors, so we will make three separate plots for each nutrient:
# Open 'pdf' graphic device. Define file name and large dimensions
pdf(file="NEON_Soils_Nutrient_Clusters.pdf", width=24, height=14)
# Set plot margins
par(mar=c(8,2,5,1), mfrow=c(3,1), xpd=NA)
# Make plots for each nutrient of interest
plotProfileDendrogram(S, d.diana.nutrients, scaling.factor = .6,
y.offset = 2, width=0.25, name='horizonName',
label='siteLabel', color='nitrogenTot',
col.label='Total Nitrogen (g/Kg)',
col.legend.cex = 1.2, n.legend=6,
col.palette=viridis::viridis(10))
plotProfileDendrogram(S, d.diana.nutrients, scaling.factor = .6,
y.offset = 2, width=0.25, name='horizonName',
label='siteLabel', color='carbonTot',
col.label='Total Carbon (g/Kg)',
col.legend.cex = 1.2, n.legend=6,
col.palette=viridis::viridis(10))
plotProfileDendrogram(S, d.diana.nutrients, scaling.factor = .6,
y.offset = 2, width=0.25, name='horizonName',
label='siteLabel', color='sulfurTot',
col.label='Total Sulfur (g/Kg)',
col.legend.cex = 1.2, n.legend=6,
col.palette=viridis::viridis(10))
# Close and save the device
dev.off()
## quartz_off_screen
## 2
This plot is not shown on this tutorial webpage, but you can view and download an example of the