Tutorial
Principal Component Analysis of AOP Hyperspectral Data in GEE
Authors: John Musinsky, Kel Markert
Last Updated: May 5, 2025
The principal components (PC) transform is a spectral rotation that takes spectrally correlated image data and outputs uncorrelated data. This can be a useful tool, especially for the high-dimensional AOP data, which is comprised of 426 bands (~380 valid bands, excluding the water vapor and noisy bands). Many of these bands may be correlated, so PCA can be a useful first step for reducing dimensionality and creating a more manageable (smaller) dataset to work with for further analysis, without reducing the essential information.
For this example, we'll use bidirectional reflectance data over the NEON site Little Rock Lake (LIRO) in Wisconsin.
Objectives
After completing this activity, you will be able to:
- Apply Principal Component Analysis (PCA) to NEON AOP hyperspectral reflectance data to reduce data dimensionality
- Create a reproducible workflow for processing high-dimensional spectral data
- Export and load PCA results as Earth Engine assets
- Compare original hyperspectral bands with PCA-transformed data
- Interpret the information content in different principal components
- Run a k-means clustering classification on the transformed dataset
- Understand some basic troubleshooting steps in case you run into errors
Requirements
- An Earth Engine account. You can sign up for an Earth Engine account here:
- An understanding of the GEE code editor and the GEE JavaScript API. See for a basic introduction.
- Optionally, complete the first three GEE tutorials in the Intro to AOP Data in Google Earth Engine Tutorial Series
- A basic understanding of dimensionality reduction and PCA concepts. If this is your first time working with PCA, we recommend reviewing this concept in the context of hyperspectral data analysis.
Additional Resources
The links below to the Earth Engine Eigen Analysis guide and KMeans documentation may provide some context as you work through this lesson.
Script 1: Computing PCA
Read in the AOP Directional Reflectance Image
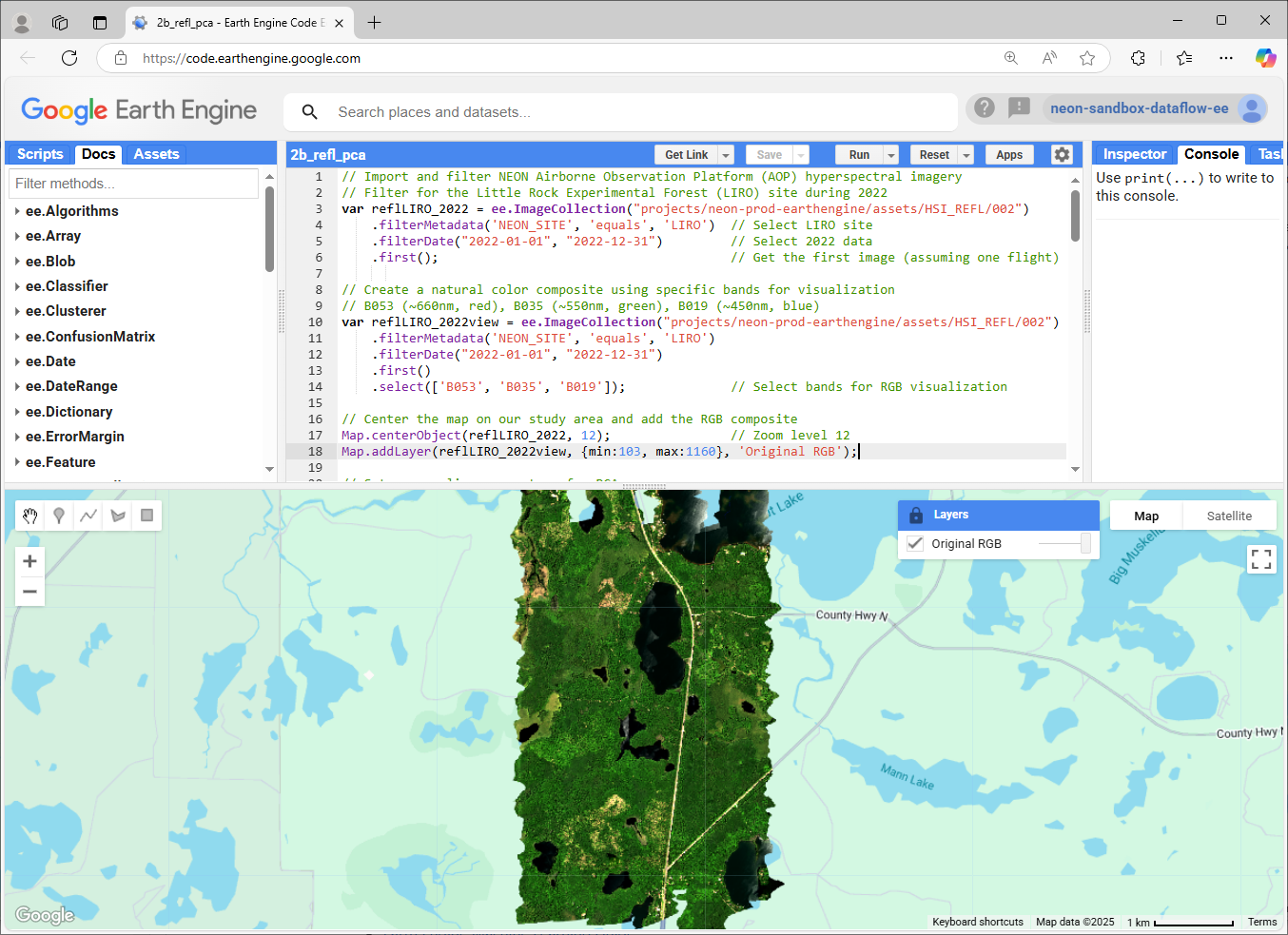
First, we'll import the hyperspectral data and create a natural color composite for visualization:
// Import and filter NEON AOP hyperspectral imagery
var reflLIRO_2022 = ee.ImageCollection("projects/neon-prod-earthengine/assets/HSI_REFL/002")
.filterMetadata('NEON_SITE', 'equals', 'LIRO') // Select LIRO site
.filterDate("2022-01-01", "2022-12-31") // Select 2022 data
.first(); // Get the first image
// Create RGB visualization using specific bands
var reflLIRO_2022view = ee.ImageCollection("projects/neon-prod-earthengine/assets/HSI_REFL/002")
.filterMetadata('NEON_SITE', 'equals', 'LIRO')
.filterDate("2022-01-01", "2022-12-31")
.first().select(['B053', 'B035', 'B019']); // Select bands for RGB visualization
// Center on the LIRO reflectance dataset
Map.centerObject(reflLIRO_2022, 12);
// Add the layer to the Map
Map.addLayer(reflLIRO_2022view, {min:103, max:1160}, 'Original RGB');
Set up Sampling for PCA
You can run PCA on an entire image, but this would be very memory intensive. Instead, we can use representative samples to compute the covariance matrix, which will provide similar results but run much more quickly. For this example, we'll collect 500 random samples. On your own you can try out different sample sizes to get a better understanding of the trade-offs.
var numberOfSamples = 500;
var sample = reflLIRO_2022.sample({
region: reflLIRO_2022.geometry(),
scale: 10,
numPixels: numberOfSamples,
seed: 1,
geometries: true
});
var samplePoints = ee.FeatureCollection(sample);
Create Helper Functions
We need two main helper functions to generate the PCA. The first function generates band names and the second calculates the PCA.
// Function to generate names for the principal component bands
// Example: PC1, PC2, PC3, etc.
function getNewBandNames(prefix, num) {
return ee.List.sequence(1, num).map(function(i) {
return ee.String(prefix).cat(ee.Number(i).int().format());
});
}
/// Function to perform Principal Component Analysis
function calcImagePca(image, numComponents, samplePoints) {
// Convert the image into an array for matrix operations
var arrayImage = image.toArray();
var region = samplePoints.geometry();
// Calculate mean values for each band
var meanDict = image.reduceRegion({
reducer: ee.Reducer.mean(),
geometry: region,
scale: 10,
maxPixels: 1e13,
bestEffort: true,
tileScale: 16 // Parameter to prevent computation timeout
});
// Center the data by subtracting the mean
var meanImage = ee.Image.constant(meanDict.values(image.bandNames()));
var meanArray = meanImage.toArray().arrayRepeat(0, 1);
var meanCentered = arrayImage.subtract(meanArray);
// Calculate the covariance matrix
var covar = meanCentered.reduceRegion({
reducer: ee.Reducer.centeredCovariance(),
geometry: region,
scale: 10,
maxPixels: 1e13,
bestEffort: true,
tileScale: 16
});
// Compute eigenvalues and eigenvectors
var covarArray = ee.Array(covar.get('array'));
var eigens = covarArray.eigen();
var eigenVectors = eigens.slice(1, 1); // Extract eigenvectors
// Project the mean-centered data onto the eigenvectors
var principalComponents = ee.Image(eigenVectors)
.matrixMultiply(meanCentered.toArray(1));
// Return the desired number of components
return principalComponents
.arrayProject([0]) // Project the array to 2D
.arraySlice(0, 0, numComponents); // Select the first n components
}
Apply PCA and Export Results
Now we'll apply the PCA to the LIRO hyperspectral image and export the results. Change the assetId tag below to point to your cloud project. This exporting step can take several minutes to complete, and longer if you are using a larger AOP image.
// Apply PCA to the hyperspectral image
var numComponents = 5; // Number of components to retain
var pcaImage = calcImagePca(reflLIRO_2022, numComponents, samplePoints);
var bandNames = getNewBandNames('PC', numComponents);
var finalPcaImage = pcaImage.arrayFlatten([bandNames]); // Convert to regular image
// Export the PCA results to Earth Engine Assets, changing the assetId so that it points to your cloud project
// This step may take around 10 minutes to complete
Export.image.toAsset({
image: finalPcaImage,
description: 'PCA_LIRO_2022',
assetId: 'projects/neon-sandbox-dataflow-ee/assets/PCA_LIRO_2022', // change this to your cloud project
scale: 1, // Output resolution in meters
maxPixels: 1e13 // Increase max pixels for large exports
});
Script 2: Visualizing PCA Results and K-Means Classification
Now that we've exported the 5 principal components, let's try to understand the results. We will then apply a K-means clustering algorithm to carry out a basic classification, to show that we can run a similar analysis on the dimensionally-reduced dataset.
Part 1: Visualizing PCA Results
After the export completes, run this second script to visualize the Principal Components:
// Load the original hyperspectral image, selecting bands for RGB visualization
// B053 (~660nm, red), B035 (~550nm, green), B019 (~450nm, blue)
var reflLIRO_2022view = ee.ImageCollection("projects/neon-prod-earthengine/assets/HSI_REFL/002")
.filterMetadata('NEON_SITE', 'equals', 'LIRO') // Select LIRO experimental forest site
.filterDate("2022-01-01", "2022-12-31") // Select 2022 data
.first() // Get first (and likely only) image
.select(['B053', 'B035', 'B019']); // Select bands for natural color display
// Load the pre-computed PCA results from Earth Engine Assets
// This asset was created by Script 1 and contains the first 5 principal components
var pcaAsset = ee.Image('projects/neon-sandbox-dataflow-ee/assets/PCA_LIRO_2022');
print("PCA image - top 5 PCA bands", pcaAsset)
// Center the map on our study area
// Zoom level 12 provides a good overview of the LIRO site
Map.centerObject(reflLIRO_2022view, 13);
// Add layers to the map
// Start with the original RGB image as the base layer
Map.addLayer(reflLIRO_2022view,
{min: 103, max: 1160}, // Set visualization parameters
'Original RGB'); // Layer name in the Layer Manager
// Pull in the palettes package and create a spectral color palette for visualization
var palettes = require('users/gena/packages:palettes');
var pc1_palette = palettes.colorbrewer.Spectral[9]
// Add the first and second principal components as layers
// PC1 typically contains the most variance/information from the original bands
Map.addLayer(pcaAsset,
{bands: ['PC1'], // Display the first component
min: -7000, max: 40000, // Set stretch values for good contrast
palette: pc1_palette,}, // Add a the pc1_palette
'PC1'); // Layer name
Map.addLayer(pcaAsset,
{bands: ['PC2'], // Display the second component
min: -7000, max: 40000, // Set stretch values for good contrast
palette: pc1_palette,}, // Add a the pc1_palette
'PC1'); // Layer name
// Note: You can toggle layer visibility and adjust transparency
// using the Layer Manager panel in the upper right of the map
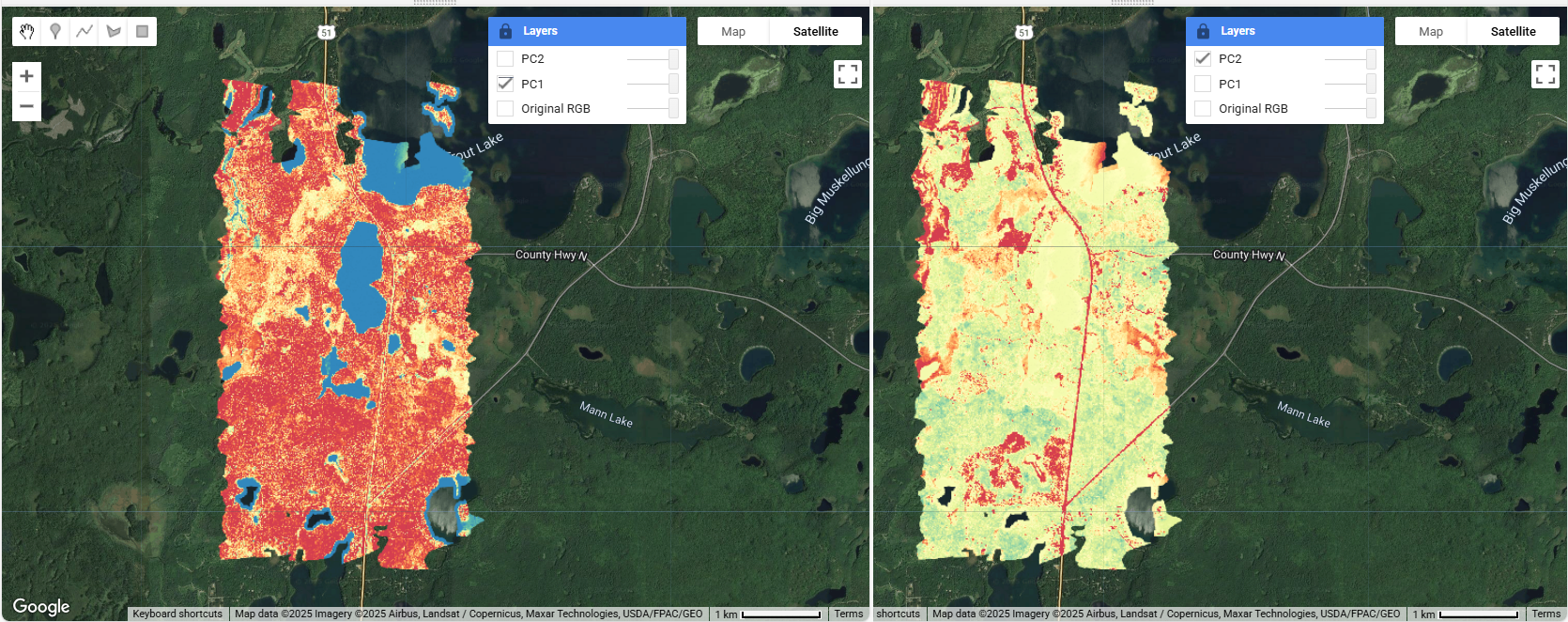
Interpreting Principal Components
- PC1: Usually represents overall brightness/albedo variations (typically 90%+ of variance)
- PC2: Often highlights vegetation vs. non-vegetation contrasts
- PC3: May reveal subtle features not visible in original bands
On your own:
- Compare PC1 with the original RGB image to understand major landscape features
- Add PC3 to the map as a layer, and look for patterns in PC2 and PC3 that might reveal hidden information
- Consider how different PCs might be useful for your specific research questions
Part 2: K-Means Classification
Now that we've run the PCA, we can use the condensed 5-band version of the data instead of the full hyperspectral dataset for further analysis. In this example, we'll show how to run a k-means clustering analysis. Kmeans is a popular unsupervized clustering algorithm for carrying out classification when you don't have training data. The code below shows how to do this:
// Create training dataset from PCA results
var training = pcaAsset.sample({
region: pcaAsset.geometry(),
scale: 10,
numPixels: 5000,
seed: 123
});
// Function to perform clustering with different numbers of clusters
function performClustering(numClusters) {
// Train the clusterer
var clusterer = ee.Clusterer.wekaKMeans({
nClusters: numClusters,
seed: 123
}).train(training);
// Cluster the PCA image
var clustered = pcaAsset.cluster(clusterer);
// Add clustered image to map
Map.addLayer(clustered.randomVisualizer(), {},
'Clusters (k=' + numClusters + ')');
return clustered;
}
// Try different numbers of clusters
var clusters5 = performClustering(5);
var clusters7 = performClustering(7);
var clusters10 = performClustering(10);
// Optional: Calculate and export cluster statistics
var calculateClusterStats = function(clusteredImage, numClusters) {
// Calculate area per cluster
var areaImage = ee.Image.pixelArea().addBands(clusteredImage);
var areas = areaImage.reduceRegion({
reducer: ee.Reducer.sum().group({
groupField: 1,
groupName: 'cluster',
}),
geometry: pcaAsset.geometry(),
scale: 10,
maxPixels: 1e13
});
print('Cluster areas (m虏) for k=' + numClusters, areas);
};
calculateClusterStats(clusters5, 5);
calculateClusterStats(clusters7, 7);
calculateClusterStats(clusters10, 10);
// Uncomment to export clustered results to your Google Drive, if desired
// Export.image.toDrive({
// image: clusters5,
// description: 'LIRO_PCA_Clusters_k5',
// scale: 5,
// maxPixels: 1e13
// });
Recap
In this lesson you:
- Created a workflow that handles large datasets efficiently
- Learned how to implement Pricipal Component Analysis (PCA) on hyperspectral data in GEE
- Visualized and exported transformed data
- Gained experience interpreting PCA results
- Gained experience running k-Means clustering and interpreting results
Troubleshooting Tips
If you run into any code errors or issues with the code, we suggest following the tips below. Errors will show up in Red in the Console, and adding print statements in the code can help you find out where the errors are occuring, if it's not obvious from the message.
Memory Limits
If you encounter "User memory limit exceeded" in the calcImagePca function, try the following:
- Increase the
scaleparameter (try 20 or 30) - Increase
tileScaleup to 16 - Reduce the region size if possible
Export Issues
If the export fails in Export.image.toAsset (at the end of Script 1):
- Verify the asset path is valid
- Check project permissions
- Try increasing
maxPixels - Allow sufficient time for processing (exports can take 10 minutes to over an hour, for some of the larger sites)
Visualization Problems
If the PCA results don't display:
- Verify the export completed successfully
- Check the asset path in Script 2
- Adjust the visualization parameters
- Try displaying one band at a time
Acknowledgment and References
Thanks to Kel Markert (Google Cloud Geographer), for his help in developing the representative sampling code as a memory-efficient option to compute PCA.
This tutorial was made with help from AI, which pulled from the following sources:
Cawse-Nicholson, K., Townsend, P.A., Schimel, D. et al. (2021). NASA's surface biology and geology designated observable: A perspective on surface imaging algorithms. Remote Sensing of Environment, 257, 112349.
- Paper discussing hyperspectral imaging algorithms including preprocessing workflows
Datt, B., McVicar, T. R., Van Niel, T. G., Jupp, D. L., & Pearlman, J. S. (2003). Preprocessing EO-1 Hyperion hyperspectral data to support the application of agricultural indexes. IEEE Transactions on Geoscience and Remote Sensing, 41(6), 1246-1259.
- Discusses preprocessing steps for hyperspectral data
Deschamps, B., McNairn, H., Shang, J., & Jiao, X. (2012). Towards operational radar-only crop type classification: comparison of a traditional decision tree with a random forest classifier. Canadian Journal of Remote Sensing, 38(1), 60-68.
- Application of PCA and clustering for classification
Green, A. A., Berman, M., Switzer, P., & Craig, M. D. (1988). A transformation for ordering multispectral data in terms of image quality with implications for noise removal. IEEE Transactions on Geoscience and Remote Sensing, 26(1), 65-74.
- Classic paper introducing PCA for dimensionality reduction in remote sensing
National Ecological Observatory Network. (2023). Data Tutorial: Introduction to Hyperspectral Remote Sensing Data. /resources/learning-hub/tutorials/hsi-hdf5-r
- NEON's introduction to hyperspectral data
Plaza, A., Benediktsson, J. A., Boardman, J. W., Brazile, J., Bruzzone, L., Camps-Valls, G., ... & Trianni, G. (2009). Recent advances in techniques for hyperspectral image processing. Remote Sensing of Environment, 113, S110-S122.
- Comprehensive review of hyperspectral processing techniques including PCA
Wang, J., & Chang, C. I. (2006). Independent component analysis-based dimensionality reduction with applications in hyperspectral image analysis. IEEE Transactions on Geoscience and Remote Sensing, 44(6), 1586-1600.
- Comparison of PCA with other dimensionality reduction techniques